Isoelectric Point Calculator 2.0
Prediction of isoelectric point and pKa dissociation constants using deep learning
Feature selection procedure
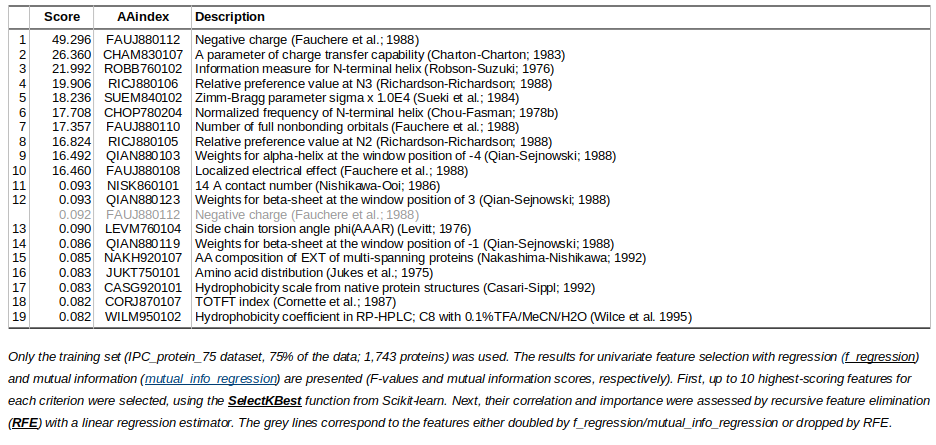
One of the first steps in machine learning is the identification of informative features. In this project apart from the amino acid sequence, one can identify additional features to boost the prediction accuracy. For this purpose, AAindex database has been used.The most informative AAindex features for protein isoelectric point prediction (IPC_protein_75 dataset)
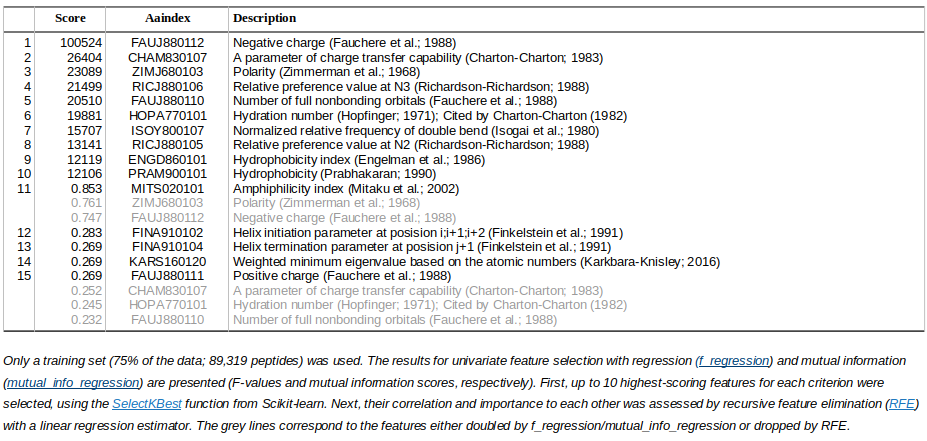
The most informative AAindex features for peptide isoelectric point prediction (IPC2_peptide_75 dataset)
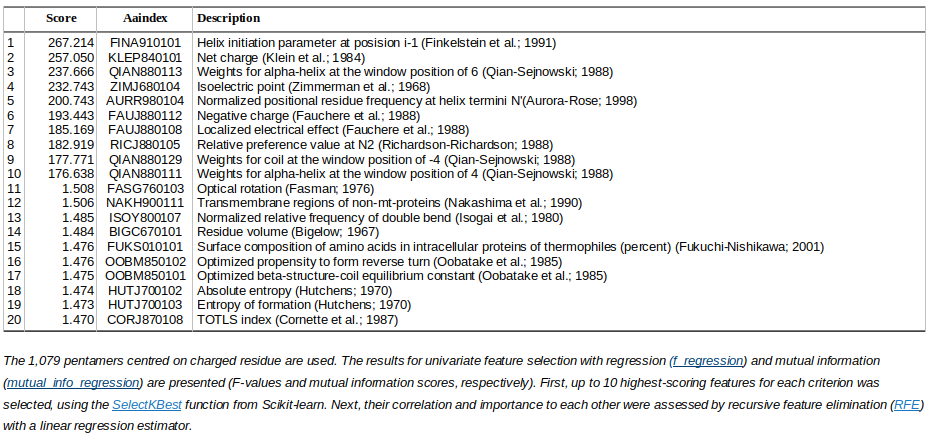
The most informative AAindex features for pKa prediction (IPC2_pKa_75 dataset)
Machine learning
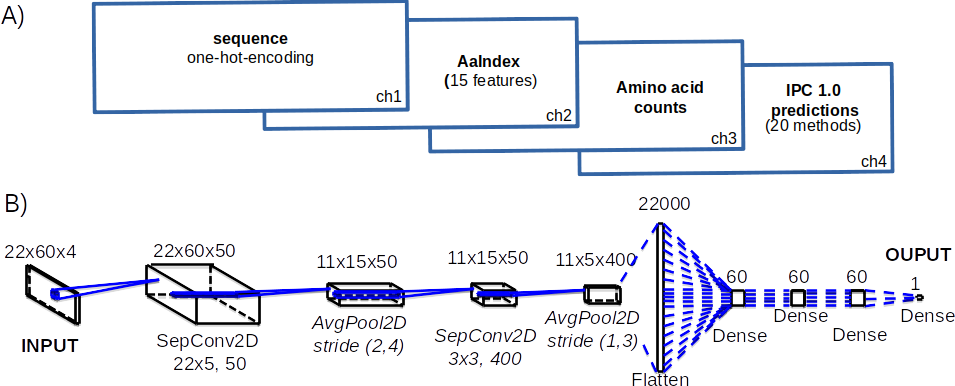
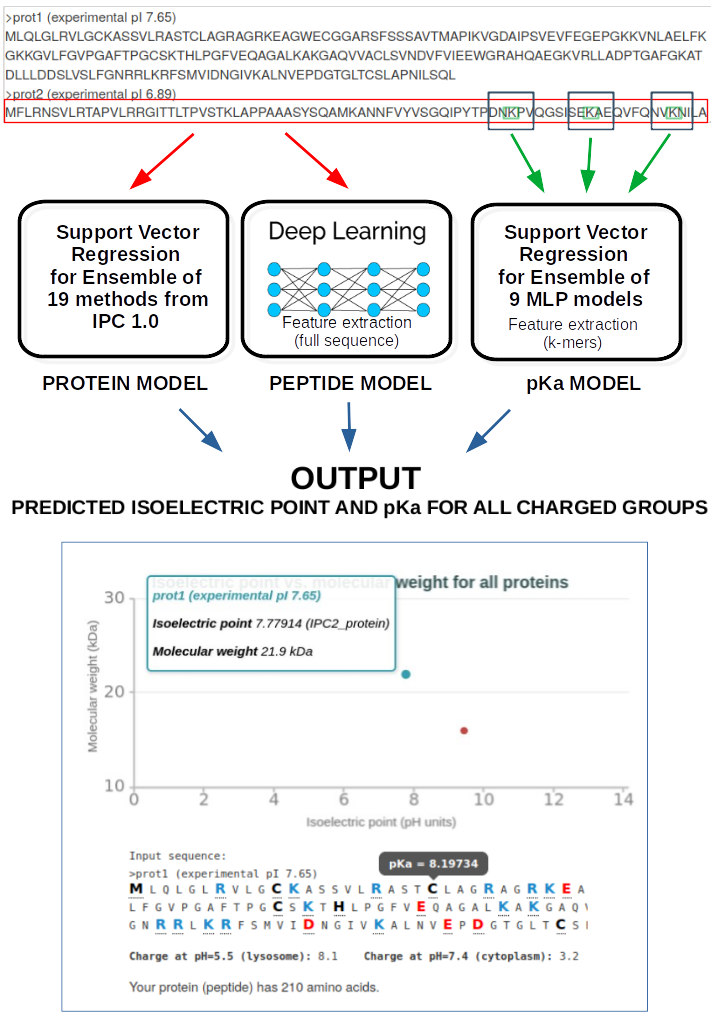
The amino acid sequences (one-hot-encoding) plus extra features (AAindex) are converted into the input that can be used in machine learning supervised training. The number of machine learning algorithms had been tested (deep learning, support vector regresion, differential evolution, basin-hopping). To avoid the overfitting 10-fold cross-validation and the dropout had been used.In a nutshell, we used:
Or to be more exact:
where for instance deep learning model for peptide (IPC2.peptide.Conv2D) looks like that: