Isoelectric Point Calculator 2.0
Prediction of isoelectric point and pKa dissociation constants using deep learning
Isoelectric point
Isoelectric point (pI) is a pH in which the net charge of the protein is zero. In case of proteins, isoelectric point mostly depends on seven charged amino acids: glutamate (δ-carboxyl group), aspartate (ß-carboxyl group), cysteine (thiol group), tyrosine (phenyl group), histidine (imidazole side chains), lysine (ε-ammonium group) and arginine (guanidinium group). Additionally, one should take into account the charge of protein terminal groups (NH2 i COOH). Each of them has its unique acid dissociation constant referred to as pK.Moreover, the net charge of the protein is in tight relation with the solution (buffer) pH.
Isoelectric point prediction
The simplest way to predict pI is to use the Henderson-Hasselbach equation to calculate protein charge in a certain pH. For more details see IPC 1.0 theory section.Yet, this is a very crude way to do so, thus in IPC 2.0, we use deep learning. For more details see the Algorithms section.
Isoelectric point importance
Proteins are the macromolecules build from 20 amino acids out of which 7 carry a charge. Additionally, at the ends, there are COO- and NH+ groups. Therefore, if those groups are exposed to the environment the whole molecule obtains certain charge which depends also on the pH of the environment. In consequence, the net charge of the protein is different at different pH. Which has important consequences for protein precipitation, X-ray crystallisation, and crystallization and few other molecular techniques. Let's analyze this on some simple example: |
Green Fluorescent Protein (Aequorea victoria) PDB: 1B9CASKGEELFTGVVPILVELDGDVNGHKFSVSGEGEGDATYGKLTLKFICTTGKLPVPWPTLVTTFTYGVQCFSRYPDHMKQ HDFFKSAMPEGYVQERTISFKDDGNYKTRAEVKFEGDTLVNRIELKGIDFKEDGNILGHKLEYNYNSHNVYITADKQKNG IKANFKIRHNIEDGSVQLADHYQQNTPIGDGPVLLPDNHYLSTQSALSKDPNEKRDHMVLLEFVTAAGITHGMDELYKWell studied protein with the molecular weight of 26.9 kDa (238 aa) and isoelectric point of ≈5.8. |
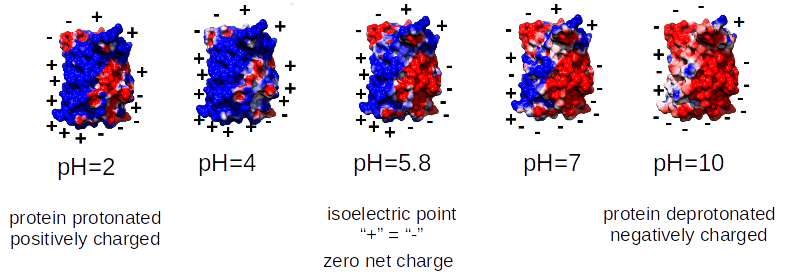
Now, you may ask for the repercussions of that:
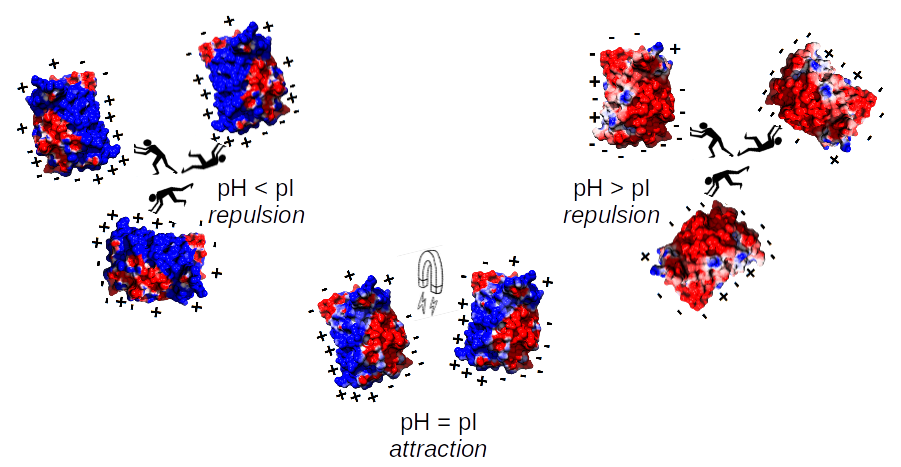
In the environment with pH similar to the isoelectric point of the protein, the molecules retain regions of negative and positive charge on their surface, which results in the tendency to aggregate. In consequence, proteins in such situations precipitate, you can not obtain protein crystals for X-ray, etc. This is the main reason you want to know a priori isoelectric point. Obviously, this has its own biological consequences as well. Simply, the organisms avoid the high expression of proteins with particular pH in given compartments (pI of the protein similar to the pH of the cellular compartment) to dodge unfavorable interactions and the aggregation.
Some pKa disociation constants
| Amino acid | NH2 | COOH | C | D | E | H | K | R | Y |
| IPC2_protein | 5.779 | 6.065 | 7.890 | 3.766 | 4.497 | 5.492 | 9.247 | 10.223 | 11.491 |
| IPC2_peptide | 7.947 | 2.977 | 9.439 | 3.969 | 4.507 | 6.439 | 8.165 | 11.493 | 9.153 |
| IPC_protein | 9.094 | 2.869 | 7.555 | 3.872 | 4.412 | 5.637 | 9.052 | 11.84 | 10.85 |
| IPC_peptide | 9.564 | 2.383 | 8.297 | 3.887 | 4.317 | 6.018 | 10.517 | 12.503 | 10.071 |
| EMBOSS | 8.6 | 3.6 | 8.5 | 3.9 | 4.1 | 6.5 | 10.8 | 12.5 | 10.1 |
| DTASelect | 8.0 | 3.1 | 8.5 | 4.4 | 4.4 | 6.5 | 10.0 | 12.0 | 10.0 |
| Solomon | 9.6 | 2.4 | 8.3 | 3.9 | 4.3 | 6.0 | 10.5 | 12.5 | 10.1 |
| Sillero | 8.2 | 3.2 | 9.0 | 4.0 | 4.5 | 6.4 | 10.4 | 12.0 | 10.0 |
| Rodwell | 8.0 | 3.1 | 8.33 | 3.68 | 4.25 | 6.0 | 11.5 | 11.5 | 10.07 |
| Patrickios | 11.2 | 4.2 | - | 4.2 | 4.2 | - | 11.2 | 11.2 | - |
| Wikipedia | 8.2 | 3.65 | 8.18 | 3.9 | 4.07 | 6.04 | 10.54 | 12.48 | 10.46 |
| Lehninger | 9.69 | 2.34 | 8.33 | 3.86 | 4.25 | 6.0 | 10.5 | 12.4 | 10.0 |
| Grimsley1 | 7.7 | 3.3 | 6.8 | 3.5 | 4.2 | 6.6 | 10.5 | 12.04 | 10.3 |
| Toseland | 8.71 | 3.19 | 6.87 | 3.6 | 4.29 | 6.33 | 10.45 | 12.0 | 9.61 |
| Thurlkill | 8.0 | 3.67 | 8.55 | 3.67 | 4.25 | 6.54 | 10.4 | 12.0 | 9.84 |
| Nozaki_Tanford | 7.5 | 3.8 | 9.5 | 4.0 | 4.4 | 6.3 | 10.4 | 12.0 | 9.6 |
| Dawson2 | 8.2 | 3.2 | 8.3 | 3.9 | 4.3 | 6 | 10.5 | 12.0 | 10.0 |
| Bjellqvist3 | 7.5 | 3.55 | 9.0 | 4.05 | 4.45 | 5.98 | 10.0 | 12.0 | 10.0 |
1 Arg was not included in the study and the average pK from all other scales was taken
2 NH2 and COOH were not included in the study and they were taken from Sillero
3 Bjellqvist model include also different pK values for terminal residues
Contact: Lukasz P. Kozlowski